References
Liu, W., Wang, Z., You, R. et al. PLMSearch: Protein language model powers accurate and fast sequence search for remote homology. Nat Commun 15, 2775 (2024). https://doi.org/10.1038/s41467-024-46808-5Liu, W. et al. (2025). PLMSearch and PLMAlign: Protein Language Model (PLM)-Based Homologous Protein Sequence Search and Alignment. In: KC, D.B. (eds) Large Language Models (LLMs) in Protein Bioinformatics. Methods in Molecular Biology, vol 2941. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-4623-6_14
References
Liu, W., Wang, Z., You, R. et al. PLMSearch: Protein language model powers accurate and fast sequence search for remote homology. Nat Commun 15, 2775 (2024). https://doi.org/10.1038/s41467-024-46808-5Liu, W. et al. (2025). PLMSearch and PLMAlign: Protein Language Model (PLM)-Based Homologous Protein Sequence Search and Alignment. In: KC, D.B. (eds) Large Language Models (LLMs) in Protein Bioinformatics. Methods in Molecular Biology, vol 2941. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-4623-6_14
About the server
 |
- Source code: github.com/maovshao/PLMAlign
Also, PLMSearch can achieve a sensitivity close to that of state-of-the-art structure search methods, while being versatile and fast based on sequences only.
- Source code: github.com/maovshao/PLMSearch
- Webserver: dmiip.sjtu.edu.cn/PLMSearch
Note: If you have needs beyond this server, send your whole input file to us at maovshao@gmail.com for help.
Tutorial
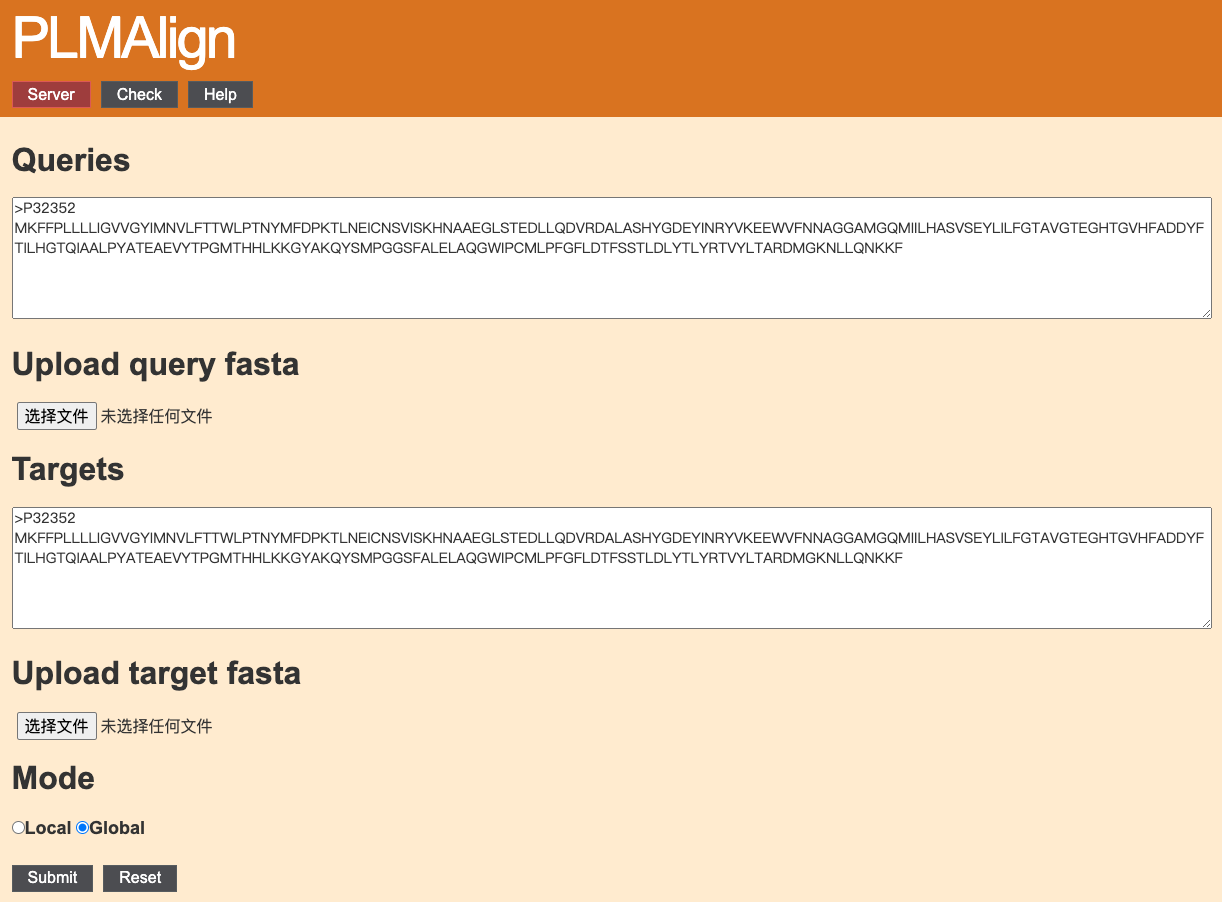 |
1. Input query and target protein sequence(s)
The sequences can either be typed directly into the text area, or uploaded from a file. If both, the server will only consider the sequence(s) in the uploaded file.Only the FASTA format is acceptable. All sequences have to be amino acids specified in a single letter code (ACDEFGHIKLMNPQRSTVWYVBZX*).
2. Submit
Press the "Submit" button to process the alignment. We will provide you a Job ID and a web link to the results.Use the "Reset" botton to clear all the inputs.
Obtain your results at any time! Just "Check" your Job ID.
Interpreting the result
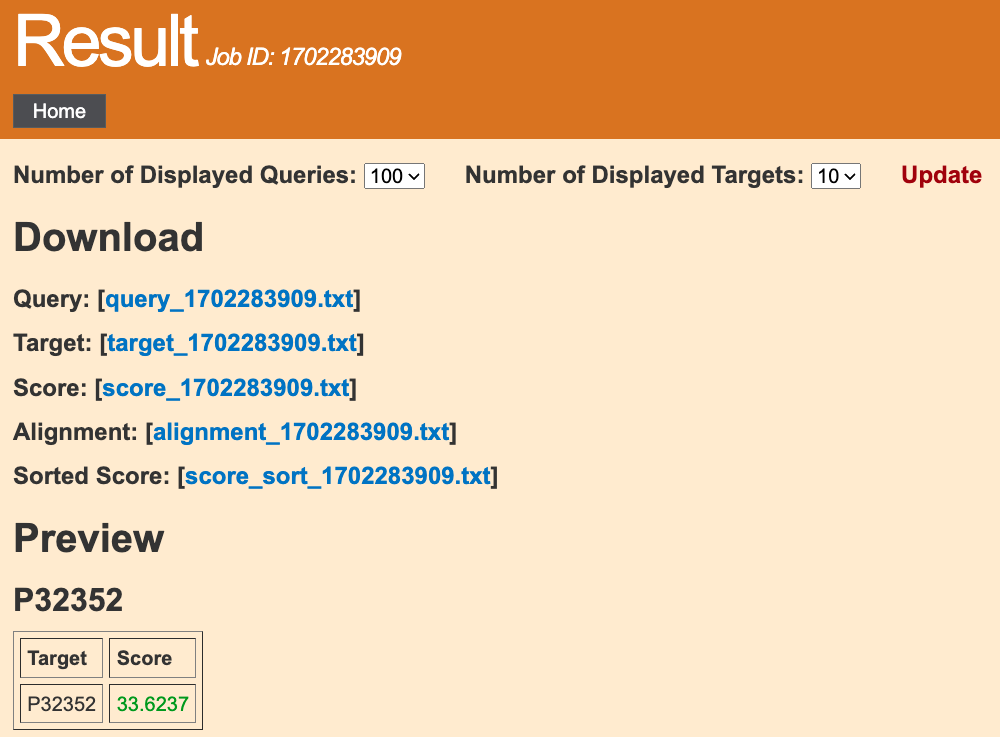 |
1. Download
Query: All your query proteins in the Fasta format.Target: All your target proteins in the Fasta format.
Score: The query-target pairs and their score.
Alignment: Sequence alignment results of query-target pairs.
Sorted Score: Consistent with Score, but the protein pairs are sorted from high to low scores.
Limitation
Query: The maximum supported number of query proteins for each submission is 100. If the number of proteins in your job exceeds, just divide them to separated jobs and submit them.2. Preview
Each table is presented for a query protein.Each row contains a target protein, arranged in descending order of score.
Reference score
Researchers often wonder what the approximate score is for homologous protein pairs. Here, we answer this question by calculating the posterior probability for proteins at certain score having the same or different folds. The same or different folds are defined by SCOP.The posterior probability of proteins with a given score being in the same fold or different fold are shown below.
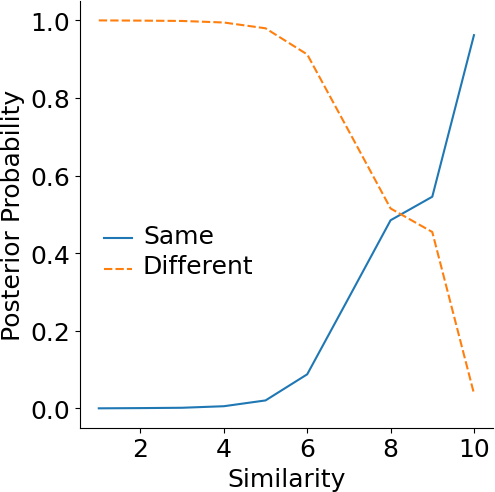 |
| Score | 3.0 | 5.0 | 7.0 | 9.0 | 9.5 |
| Posterior probability (same fold) | 0.14% | 2.03% | 28.57% | 54.54% | 75.00% |
| Posterior probability (different fold) | 99.85% | 97.96% | 71.42% | 45.45% | 25.00% |
Contact us
Please contact Shanfeng Zhu (zhusf@fudan.edu.cn) or Wei Liu (maovshao@gmail.com), if you have any question or comment about the server.User's feedback is quite important to us.
References
Liu, W., Wang, Z., You, R. et al. PLMSearch: Protein language model powers accurate and fast sequence search for remote homology. Nat Commun 15, 2775 (2024). https://doi.org/10.1038/s41467-024-46808-5Liu, W. et al. (2025). PLMSearch and PLMAlign: Protein Language Model (PLM)-Based Homologous Protein Sequence Search and Alignment. In: KC, D.B. (eds) Large Language Models (LLMs) in Protein Bioinformatics. Methods in Molecular Biology, vol 2941. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-4623-6_14