Motivation: Automated function prediction (AFP) of proteins is of great significance for elucidating the molecular mechanism of life. As one of the state-of-the-art AFP methods, NetGO 2.0 [3]integrates multi-source information to improve the performance. However, it mainly utilizes the proteins with experimentally supported functional annotations, and thus ignores valuable information from a vast number of unannotated proteins. Recently, protein language models have been proposed to learn informative representations from protein sequences only based on self-supervision[6]. Therefore, it is promising to incorporate protein language model into NetGO 2.0 for AFP.
Results: We represent each protein by Evolutionary Scale Modeling (ESM)-1b [6] embedding and use Logistic Regression (LR) to train a new model, LR-ESM, for AFP. The experimental results show that LR-ESM significantly outperformed Seq-RNN based on recurrent neural network, which is a main component of NetGO 2.0 [3]. Therefore, we develop NetGO 3.0 as an upgraded version of NetGO 2.0 to improve the performance of AFP extensively, with the replacement of Seq-RNN by LR-ESM based on protein language model.
2021.12.01: NetGO 3.0 released.
2020.09.20: NetGO 2.0 released.
2020.02.22: NetGO 1.1 released.
2018.11.20: NetGO 1.0 released.
Results: We represent each protein by Evolutionary Scale Modeling (ESM)-1b [6] embedding and use Logistic Regression (LR) to train a new model, LR-ESM, for AFP. The experimental results show that LR-ESM significantly outperformed Seq-RNN based on recurrent neural network, which is a main component of NetGO 2.0 [3]. Therefore, we develop NetGO 3.0 as an upgraded version of NetGO 2.0 to improve the performance of AFP extensively, with the replacement of Seq-RNN by LR-ESM based on protein language model.
Version History
2025.02.10: NetGO 4.0 released.2021.12.01: NetGO 3.0 released.
2020.09.20: NetGO 2.0 released.
2020.02.22: NetGO 1.1 released.
2018.11.20: NetGO 1.0 released.
References
- You R, Zhang Z, Xiong Y, et al. GOLabeler: improving sequence-based large-scale protein function prediction by learning to rank. Bioinformatics, Volume 34, Issue 14, 15 July 2018, Pages 2465–2473.
- Zhou N, Jiang Y, Bergquist T R, et al. The CAFA challenge reports improved protein function prediction and new functional annotations for hundreds of genes through experimental screens. Genome biology, 2019, 20(1): 1-23.
- Yao S, You R, Wang S, et al. NetGO 2.0: improving large-scale protein function prediction with massive sequence, text, domain, family and network information. Nucleic Acids Research, 2021;, gkab398.
- You R, Yao S, Xiong Y, et al. NetGO: improving large-scale protein function prediction with massive network information. Nucleic Acids Research, Volume 47, Issue W1, 02 July 2019, Pages W379–W387.
- You R, Huang X, Zhu S. DeepText2Go: Improving large-scale protein function prediction with deep semantic text representation. Methods, 2018, 145: 82-90.
- Rives A, Meier J, Sercu T, et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proceedings of the National Academy of Sciences, Volume 118, Issue 15. 2021.
- You R, Yao S, Mamitsuka H, et al. DeepGraphGO: graph neural network for large-scale, multispecies protein function prediction. Bioinformatics (ISMB 2021), Volume 37, Issue Supplement_1, July 2021, Pages i262–i271.
- Wang S, You R, Liu Y, et al. NetGO 3.0: Protein Language Model Improves Large-scale Functional Annotations. Genomics, Proteomics & Bioinformatics, In Press.
The Tutorial on NetGO 3.0 Predictions
NetGO 3.0, based on the previous version, improves large-scale automated function prediction (AFP) with a protein language model. If you want to redirect to the homepage of NetGO 2.0, you can click the upper bottom "NetGO 2.0". The steps to make a prediction and explanations of the meanings of the query results are detailed as follows.A. How to obtain predictions
This is a screenshot of an input page. The numbers in red refer to the below different sections. |
1. Input a protein sequence(s)
Firstlt, specify the sequence(s) you want to predict. The sequences can be typed directly into the text area or uploaded from a file using the button. If both the text area and the file uploaded contain sequences, this server will only consider the sequence(s) in the text area and the sequence(s) in the uploaded file will be ignored.Only the FASTA Format is acceptable to this server: By FASTA Format, long protein sequences in one or multiple lines, each of which begins with '>', are allowed. The lines starting with '>' are treated as identifiers of the following sequences.
All sequences must be amino acids specified in a single-letter code (ACDEFGHIKLMNPQRSTVWYVBZX*). The input processor will reject any other non-whitespace characters with a notification. Also, our input processor will check the empty sequence and the uniqueness of sequence identifiers, and a warning will be given when nucleotide-like sequences are found.
The web page also provides an example if you have trouble choosing protein suequence. Please click the "Show an example" to provide an example for NetGO 3.0. You can also click "Example File" to download an example file.
Note: To run NetGO 3.0, we strongly recommend you to provide proteins with standard UniProt identifiers. Otherwise, text information related to the input proteins will not be used.
2. Input your email address(Optional)
For this option, you can input your email address. We will send you a confirmation email after your submission. You will receive an email informing you that prediction results are available. Although it is optional, we highly recommend you use this service.After you have followed all the above steps, please press the "Submit" button to process the prediction. We will provide you with a job id and a web link to the results. The page will refresh automatically when your query results are available (If you have provided your email address, you will receive an email) notification . You can track your job status at any time by directly clicking the link or entering your job id on the "Check" page.
The time cost of making a prediction depends on the number of input data points. Usually, it will run relatively quickly. The below table on running times is for your reference.
| Protein Num | NetGO 3.0 | NetGO 2.0 |
| 1 | 5.2min | 7.3min |
| 100 | 14.5min | 16.8min |
| 200 | 18.2min | 23.5min |
| 400 | 32.4min | 42.4min |
| 1000 | 84.1min | 90.1min |
B. Interpreting the prediction output
When a prediction has been made, you can obtain your query results just like you track your job status. The following is a screenshot of a result page. The numbers in red refer to different sections of the interface.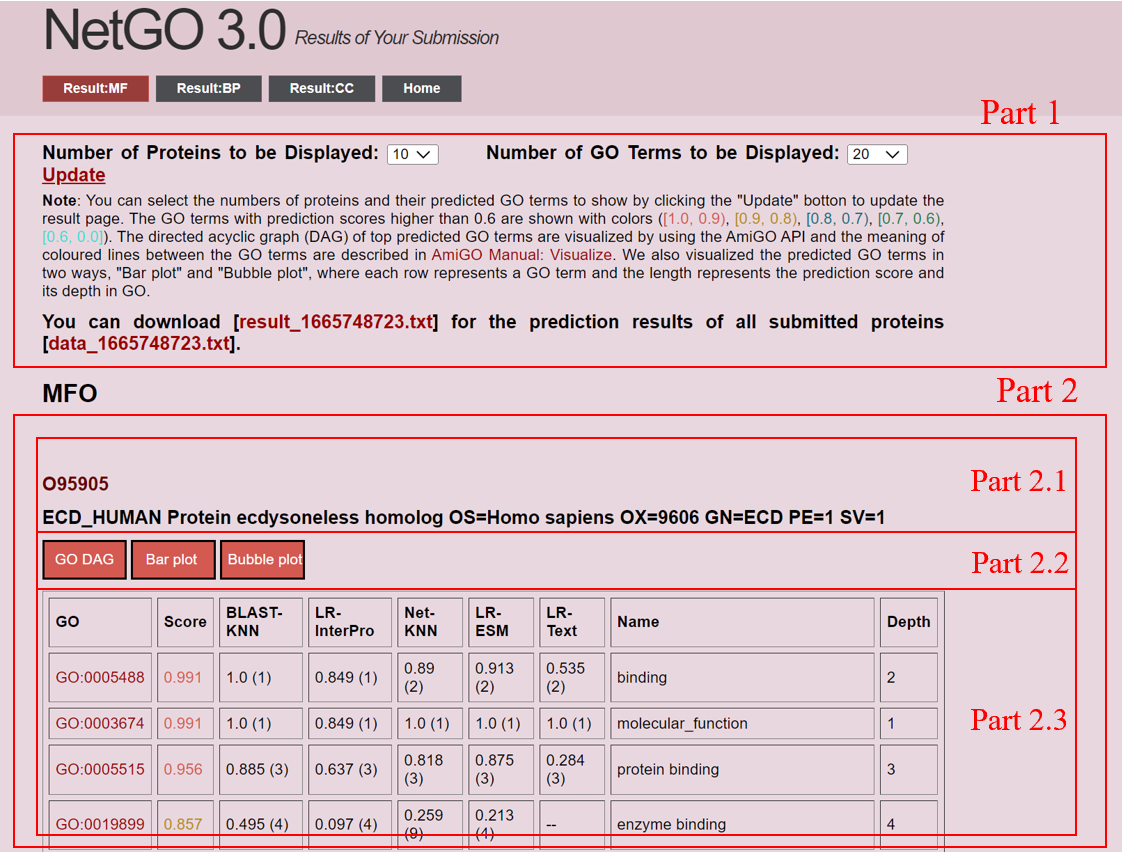 |
1. Information about results (Part 1)
We show some basic information about results and provide a link to download your results.Prediction results for a protein (Part 2)
We will only show the result of the first 10 (20 or 30, depending on the user's choice) proteins.(1) Protein name (2.1)
You can click the protein name for each protein to obtain its information in UniProt. Further, we also show the basic information for the protein in UniProt, such as alternate name, species, and genes.
(2) Prediction results shown in graph (2.2)
We visualize the prediction results in 3 ways, as shown in Part 2.2 "GO DAG", "Bar plot" and "Bubble plot".
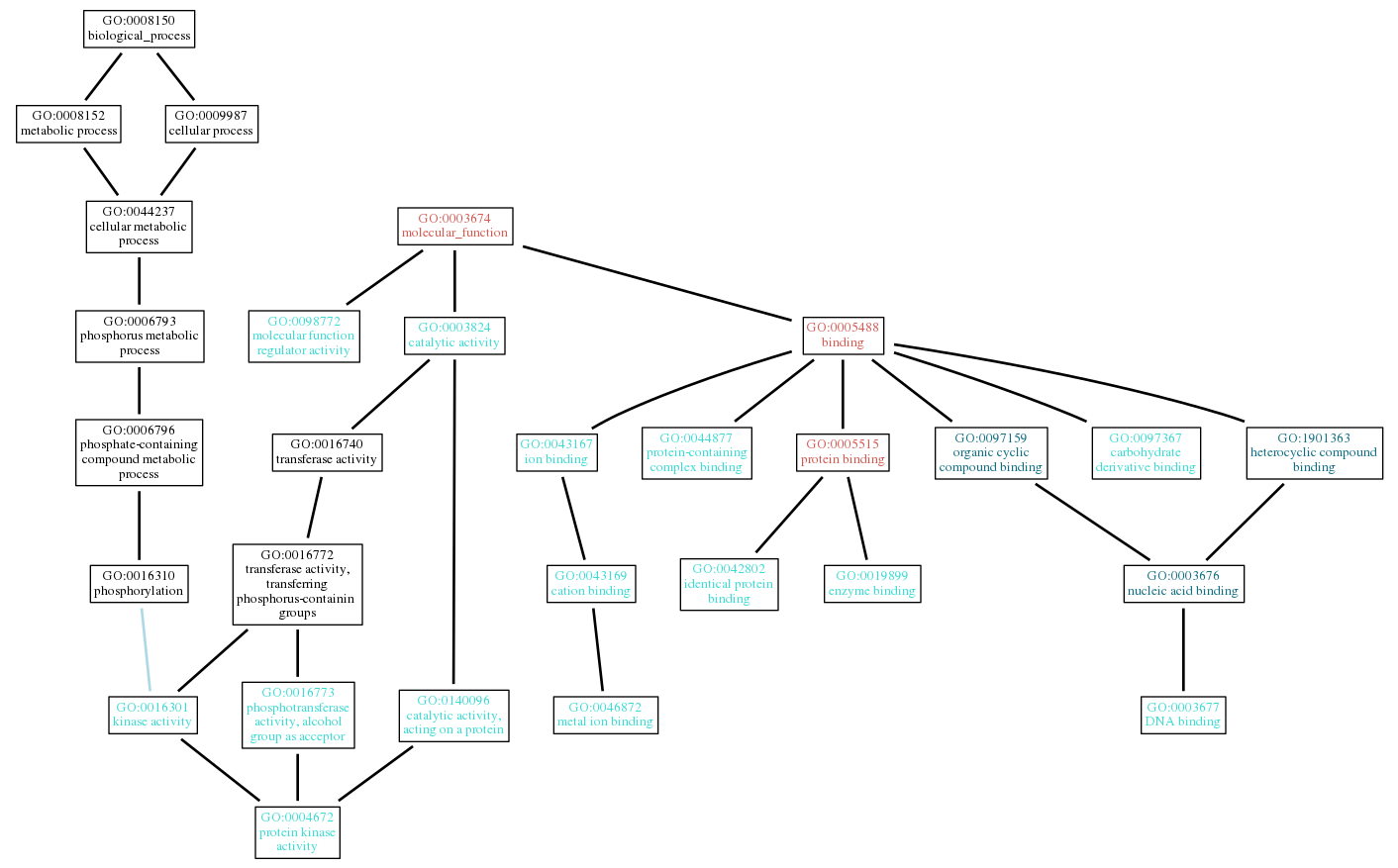
(2.2.1) GO DAG
We visualize the top m (m=20 by default, and can be set to 30, 50 or 100) predicted GO terms according to the GO directed acyclic graph (DAG) and organized all of them in a picture. Note that GO terms of high confidence (score > 0.6) will be emphasized with colors ([1.0, 0.9), [0.9, 0.8), [0.8, 0.7), [0.7, 0.6), [0.6, 0.0]). The top predictions of GO terms are visualized by using the AmiGO API. The meaning of colored lines between the GO terms are described in AmiGO Manual: Visualize, where blue color means relation "is_a", light blue color means relation "part_of", brown color represents relation "develops_from", black color stands for relation "regulates", red color indicates relation "negatively_regulates" and green color denotes relation "positively_regulates".
Note that GO terms use the "is_a" and "part_of" relationships to form directed acyclic graphs(DGA) in subontology, while the AmiGO visualization tool takes into account "develops_from", "regulates" and other relationships. So terms from other sub-ontologies or CARO and CL terms will be presented.
You can click the "GO DAG" button to show a high-resolution version.
 |
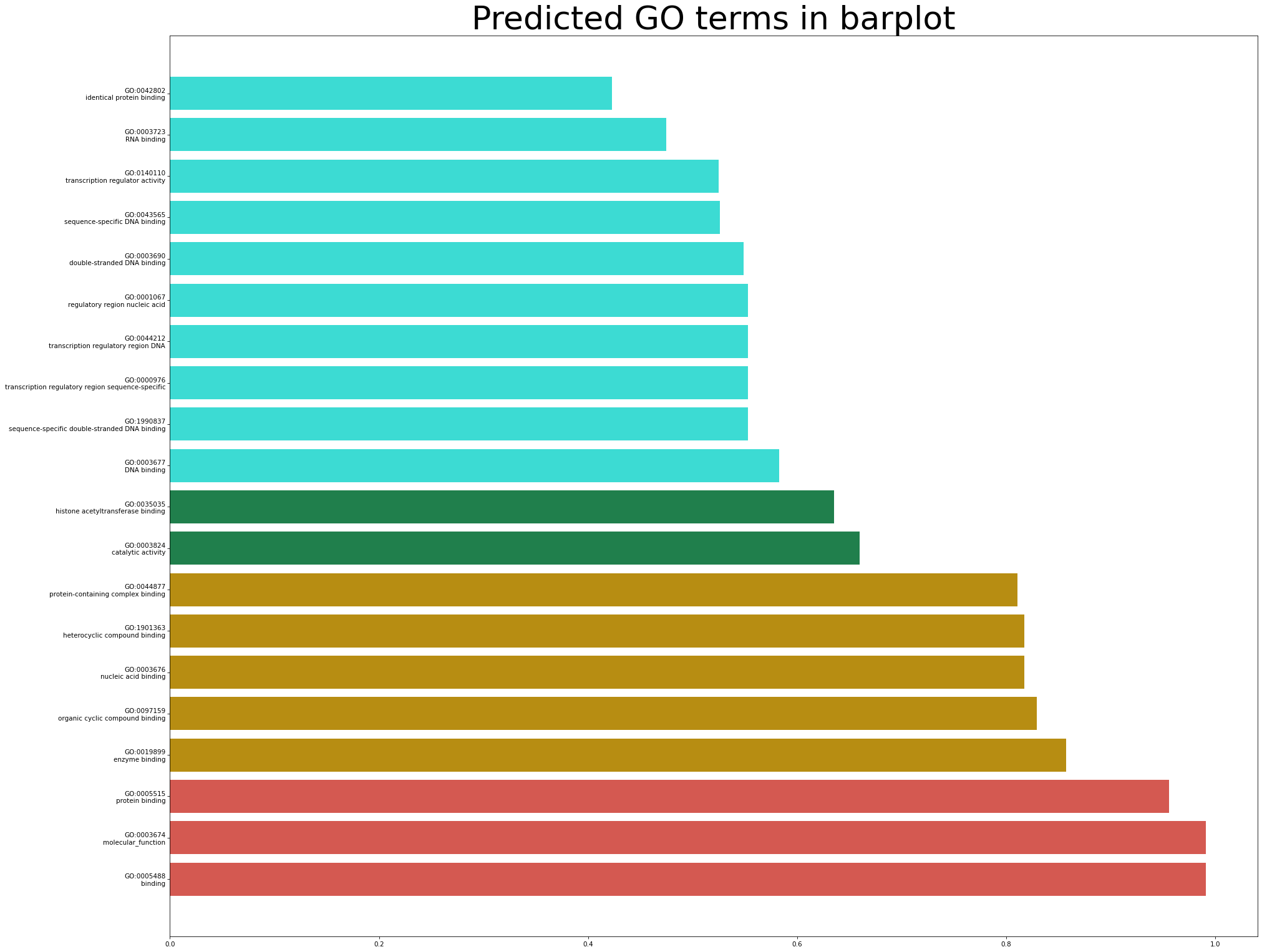
The predicted GO terms are visualized in the barplot. The GO terms and corresponding scores are depicted in the following picture. Once again the colors are associated with the confidence scores. The number of shown GO terms depends on the user's choice. You can click the button of "Bar plot" to show a high-resolution version.
 |
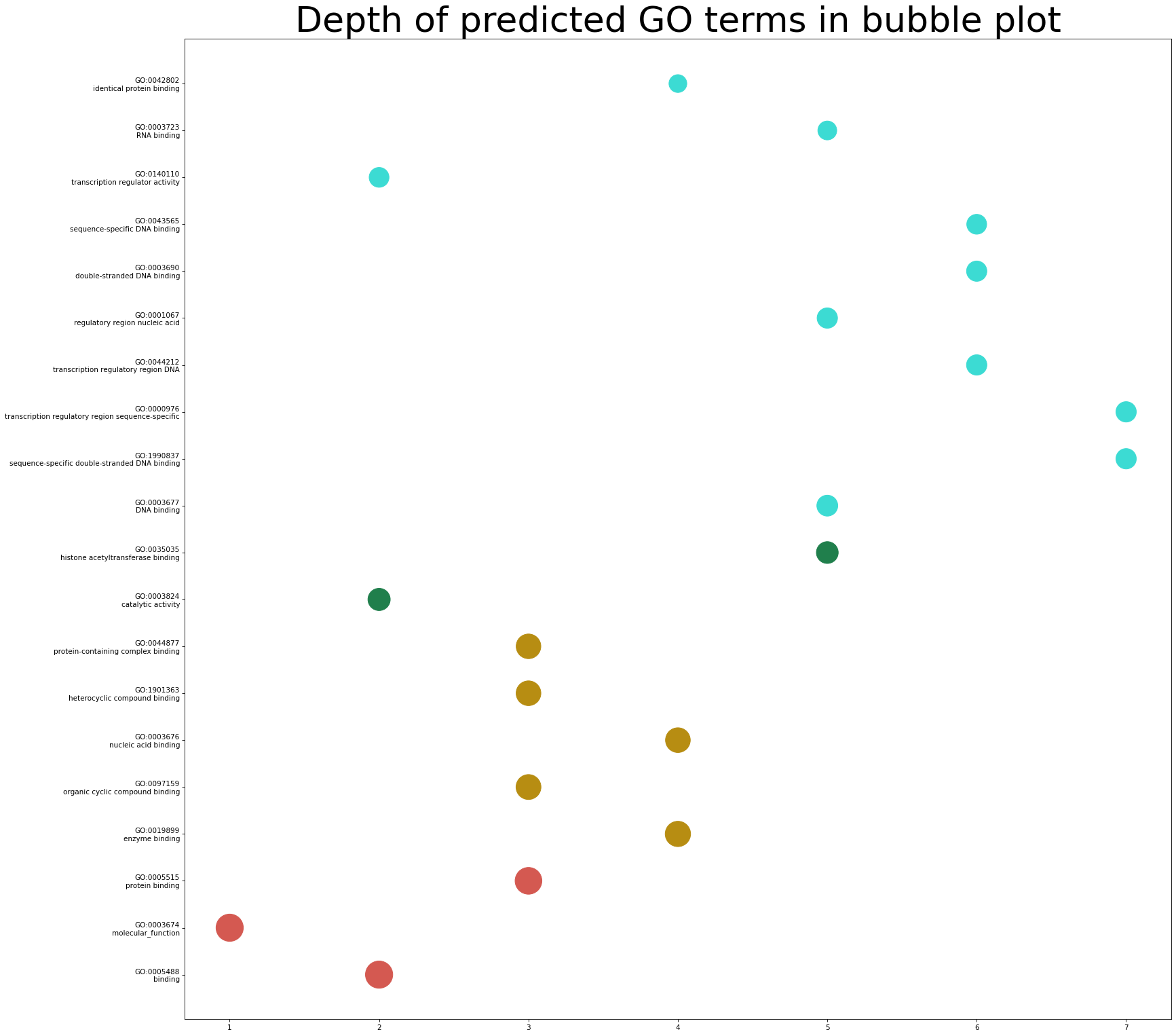
The depths of predicted GO terms are also noteworthy information as most GO terms in the deeper layers always have a more specific description of protein function. Here the size of the corresponding dot is closely related to the prediction scores. You can click the button "Bubble plot" to show a high-resolution version.
 |
The top 20 predicted GO terms are also shown in a table. The three columns list GO terms, corresponding scores, and names. The numbers between brackets are the ranks of a GO term in the prediction result of the component methods. You can click on GO terms to display their detailed information. GO terms of high confidence (score > 0.6) are also be emphasized with colors ([1.0, 0.9), [0.9, 0.8), [0.8, 0.7), [0.7, 0.6), [0.6, 0.0]).
Note that the scores, especially by the components methods, can be regarded as probabilities more or less. However, since we treat AFP as a ranking problem, NetGO 2.0 focuses more on the rank of their relative scores of different GO Terms for the same protein. The scores between proteins, components, and sub-ontologies are not directly comparable.
C. Browser compatibility
We have tested our system and browser compatibility, as shown below:| OS | Version | Chrome | Firefox | Microsoft Edge | Safari |
| Linux | Ubuntu 16 | 71.0 | 64.0 | n/a | n/a |
| MacOS | 12 | 83.0 | 84.0 | 83.0 | 13.1 |
| Windows | 10 | 87.0 | 83.0 | 87.0 | n/a |
For any scientific problems, please contact Shanfeng Zhu (zhusf@fudan.edu.cn).
If any bug occurs or for technical problems, please contact Shaojun Wang (shaojunwang20@fudan.edu.cn).
We will highly appreciate your support and kindness.
If any bug occurs or for technical problems, please contact Shaojun Wang (shaojunwang20@fudan.edu.cn).
We will highly appreciate your support and kindness.
This web server is free and open to all without a login requirement.